Submit NMR spectra to Zenodo
Submitting data to Zenodo is still an experimental feature. Please test it in the sandbox and share your feedback on https://www.nmrium.com/contact.
Visit the NMRium dataset on Zenodo to browse submitted datasets.
You may also check the NMRium dataset in the sandbox but don't forget they may deleted anytime.
Submit NMR spectra to Zenodo with direct integration into NMRium for interactive visualization and analysis. This tool enables the ultimate FAIR data submission for NMR datasets, making your research data Findable, Accessible, Interoperable, and Reusable.
About
zenodo-nmrium is a command-line tool designed to streamline the submission of NMR spectra datasets to Zenodo while leveraging the power of NMRium for interactive, web-based visualization.
Why FAIR Data Matters
This tool ensures your NMR data follows FAIR principles:
- Findable: Your data is registered and discoverable on Zenodo
- Accessible: Data is freely available on the public internet
- Interoperable: Data is stored as a zip file that contains the original NMR data that work with NMRium and other tools
- Reusable: Rich metadata and open licenses facilitate reuse by others
The Ultimate Goal
zenodo-nmrium enables a complete FAIR data workflow:
- Organize your data: Package each NMR sample in a folder or as a
.nmrium.zipfile - Store in Zenodo: Upload your data to Zenodo (sandbox for testing, production for publication)
- Interactive visualization: All data is directly accessible from Zenodo and can be viewed interactively in NMRium
- Table of contents: Navigate between multiple samples through a structured table of contents
- Community sharing: Join the NMRium community on Zenodo to discover and access all community-submitted datasets
Installation
Pre-built Binaries
Pre-built binaries are available for macOS ARM64 and Linux x64.
macOS ARM64 (Apple Silicon)
curl -o zenodo-nmrium https://www.lactame.com/lib/zenodo-nmrium/HEAD/zenodo-nmrium-darwin-arm64 && chmod 555 zenodo-nmrium
Then use it:
./zenodo-nmrium <path> [options]
Linux x64
curl -o zenodo-nmrium https://www.lactame.com/lib/zenodo-nmrium/HEAD/zenodo-nmrium-linux-x64 && chmod 555 zenodo-nmrium
Then use it:
./zenodo-nmrium <path> [options]
Other Platforms
For other platforms (Windows, macOS Intel, ARM Linux, etc.), please contact us.
Quick Start
1. Organize Your Data
Create a folder with the following structure:
- One folder or
.nmrium.zipfile per item in your menu/table of contents - Each folder/ZIP should contain all NMR data for a sample or group of samples
- One
index.ymlmetadata file at the root describing your entire dataset
Example folder structure:
my-nmr-dataset/
├── index.yml # Dataset metadata (required)
├── sample_1/ # First sample (name will be reused in the table of content)
│ ├── structure.mol # Optional: chemical structure file (molfile format)
│ ├── 12/ # Experiment number of a native Bruker folder
│ │ ├── fid
│ │ ├── acqu
│ │ ├── acqus
│ │ ├── pdata/ # Processed data folder
│ │ │ └── 1/
│ │ │ ├── proc
│ │ │ ├── procs
│ │ │ └── ...
│ │ └── ...
│ ├── 13/ # Another experiment
│ └── ...
├── sample_2.nmrium.zip # Second sample was processed using NMRium. It could contain assignment
├── sample_3/
│ ├── structure.mol # Optional: chemical structure file (molfile format)
│ └── spectrum.jdx # JCAMP-DX file (e.g., exported from Topspin using `tojdx` or from MNova with peak assignments)
└── ...
Each sample folder may optionally contain a structure.mol file (MOL format) if the chemical structure is known. This allows NMRium to display and analyze the structure alongside the NMR spectra.
2. Create the Metadata File (index.yml)
The index.yml file is required and should be placed at the root of your dataset folder. It contains metadata about your entire dataset.
Example index.yml
title: NMR spectra dataset generated with NMRium CLI
description: |-
<p>This is a dataset generated with NMRium CLI.</p>
<p>The dataset will be visible and accessible in <strong>NMRium</strong>, providing an <strong>interactive</strong> viewing experience for NMR spectra analysis.</p>
<p>This represents the <strong>ultimate FAIR data submission</strong> — ensuring that your data is Findable, Accessible, Interoperable, and Reusable.</p>
copyright: cc-by-4.0
keywords:
- NMR spectra
- FAIR data
- Nuclear Magnetic Resonance
authors:
- family_name: 'Zasso'
given_name: 'Michaël'
affiliation: 'Zakodium Sàrl, Switzerland'
orcid: '0000-0001-5295-2159'
- family_name: 'Bolaños'
given_name: 'Alejandro'
affiliation: 'Universidad del Valle, Cali, Colombia'
orcid: '0000-0001-5933-5524'
Metadata Fields
| Field | Type | Required | Description |
|---|---|---|---|
title | String | Yes | Title of your NMR dataset |
description | String (HTML) | Yes | Dataset description as a string in HTML format |
copyright | String | Yes | License identifier (default: cc-by-4.0, e.g., cc0-1.0) |
keywords | Array | No | List of keywords for dataset discoverability |
authors | Array | Yes | List of dataset authors |
Author Fields
| Field | Type | Required | Description |
|---|---|---|---|
family-name | String | Yes | Author's surname |
given-name | String | Yes | Author's first name |
affiliation | String | No | Author's institutional affiliation |
orcid | String | No | Author's ORCID identifier (format: 0000-0000-0000-0000) |
3. Get a Zenodo API Token
- Sandbox (testing): https://sandbox.zenodo.org/account/settings/applications/tokens/new/
- Production: https://zenodo.org/account/settings/applications/tokens/new/
You must enable deposit:action and deposit:write permissions when creating the token.
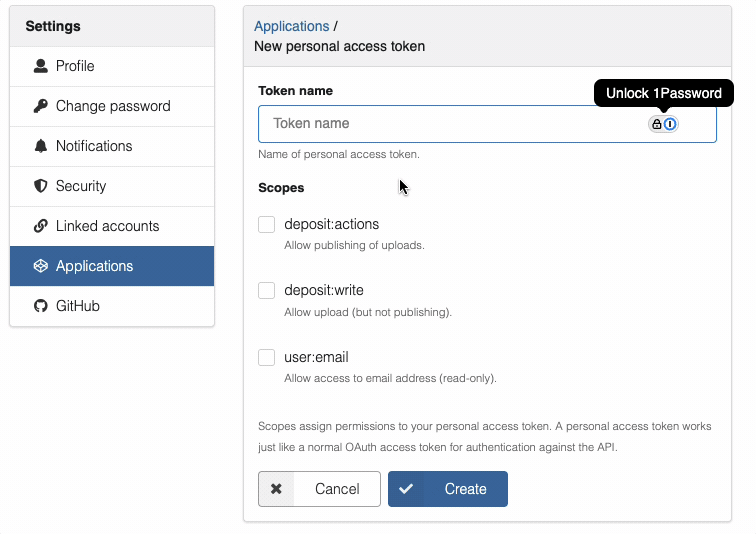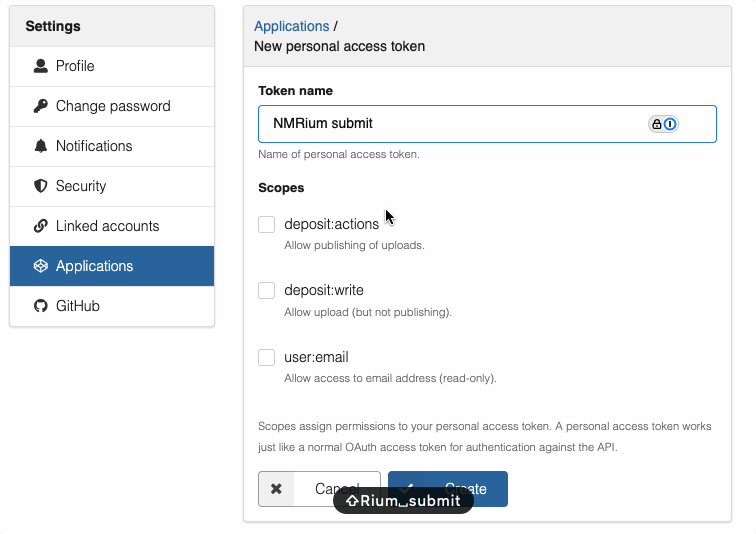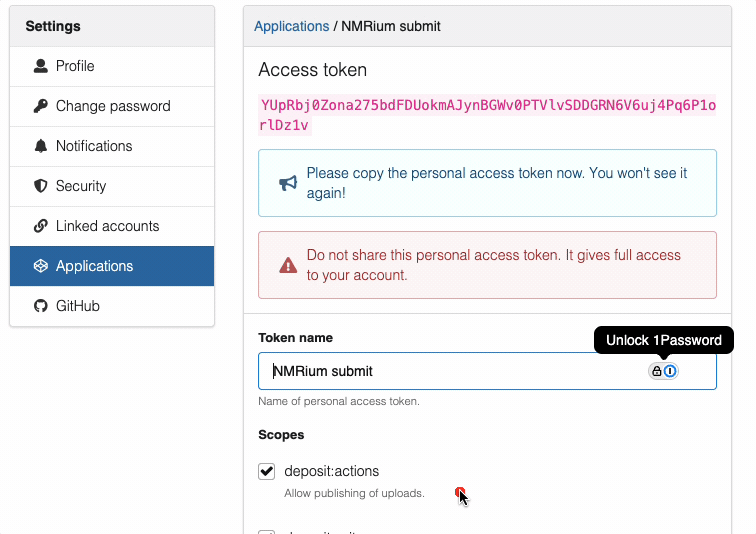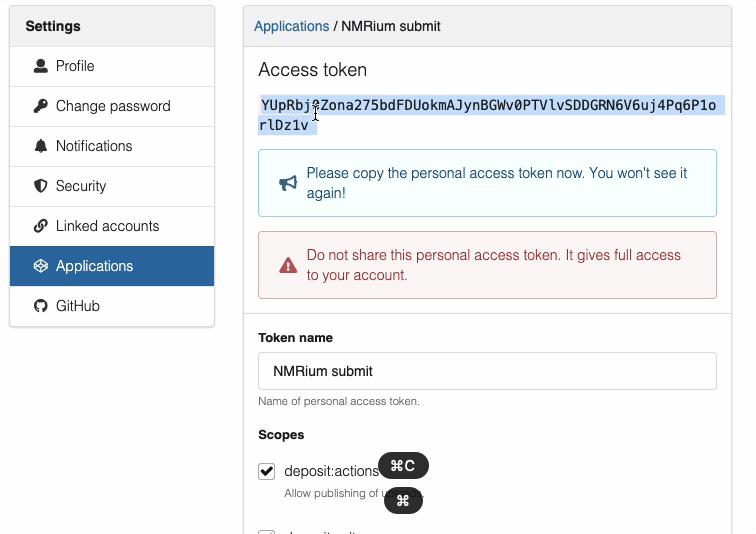
Sandbox and production tokens are always different. You will only see this token once, so save it in a secure location.
Providing the Token
You can provide your Zenodo API tokens in two ways:
Option 1: Command Line
Pass the token directly via the -t or --access-token option:
./zenodo-nmrium ./my-nmr-dataset -t YOUR_SANDBOX_TOKEN
Option 2: Environment Variables (.env file)
Store your tokens in a .env file. The tool will look for this file in one of three locations (in order):
.envin the current working directory.zenodo-nmriumin your home directory (~/.zenodo-nmrium).envin your home directory (~/.env)
Create a .env file with the following environment variables:
ACCESS_TOKEN=<ZENODO_PRODUCTION_TOKEN>
ACCESS_TOKEN_SANDBOX=<ZENODO_SANDBOX_TOKEN>
You must use two different tokens:
ACCESS_TOKENfor production submissions (-pflag)ACCESS_TOKEN_SANDBOXfor sandbox submissions (default or-sflag)
Using the wrong token for the environment will fail!
Example .env file:
# Zenodo production token
# Generated from: https://zenodo.org/account/settings/applications/tokens/new/
ACCESS_TOKEN=your_production_token_here
# Zenodo sandbox token
# Generated from: https://sandbox.zenodo.org/account/settings/applications/tokens/new/
ACCESS_TOKEN_SANDBOX=your_sandbox_token_here
When using environment variables, you can omit the -t option:
# Uses ACCESS_TOKEN_SANDBOX from .env file
./zenodo-nmrium ./my-nmr-dataset
# Uses ACCESS_TOKEN from .env file
./zenodo-nmrium ./my-nmr-dataset -p
4. Submit Your Data
Basic submission to sandbox:
./zenodo-nmrium ./my-nmr-dataset -t YOUR_SANDBOX_TOKEN
Auto-publish after submission:
./zenodo-nmrium ./my-nmr-dataset -t YOUR_SANDBOX_TOKEN -a
Submit to production:
./zenodo-nmrium ./my-nmr-dataset -t YOUR_PRODUCTION_TOKEN -p
Combine options:
./zenodo-nmrium ./my-nmr-dataset --access-token YOUR_TOKEN --production --auto-publish
Use your sandbox token for testing and your production token for final submission. These tokens are different and cannot be used interchangeably.
Command Options
Usage: zenodo-submit <path> [options]
Positional Arguments:
path Path to dataset folder with index.yml and data
Options:
-t, --access-token <token> Zenodo API access token (required)
-s, --sandbox Use sandbox environment (default)
-p, --production Use production environment
-a, --auto-publish Auto-publish after submission
-d, --data-selection <type> Data selection for NMRium visualization
Choices: ft, fid, both, preferFT, preferFID
Default: ft
-l, --log-level <level> Logger level
Choices: trace, debug, info, warn, error, fatal, silent
Default: info
-h, --help Show this help message
Option Details
Positional Argument: path (Required)
- Description: Path to your dataset folder
- Contains:
index.ymlfile and NMR data folders/ZIPs - Example:
./my-nmr-dataset,/Users/username/data/spectra
Access Token (-t, --access-token)
- Description: API token for authentication with Zenodo
- Required? Yes, either via
-toption or environment variables (ACCESS_TOKENorACCESS_TOKEN_SANDBOX) - How to get:
- Log in to Zenodo (sandbox or production)
- Go to Account → Settings → Applications → Personal access tokens
- Click "New token"
- Give it a name (e.g., "NMRium submission")
- Select the required scopes:
deposit:actionanddeposit:write - Click "Create" to generate the token
- Copy the token
- Important:
- Sandbox and production will have different tokens
- Keep tokens private and secure
- Never commit tokens to version control
- Ensure your token has
deposit:actionanddeposit:writepermissions
- Usage:
- Command line:
-t YOUR_TOKENor--access-token YOUR_TOKEN - Environment: Set
ACCESS_TOKENorACCESS_TOKEN_SANDBOXin a.envfile
- Command line:
Environment (-s, --sandbox | -p, --production)
- Default: Sandbox (if neither
-snor-pis specified, data will be submitted to sandbox) - Usage:
-sor no flag: Submit to Zenodo sandbox (test environment)-p: Submit to production environment (public, permanent)
- Important: Cannot use both
-sand-psimultaneously - Workflow: Test in sandbox first, then use
-pfor production
Sandbox vs. Production:
| Aspect | Sandbox | Production |
|---|---|---|
| Purpose | Testing and development | Final publication |
| Data retention | Cleaned periodically | Permanent |
| Public access | Limited | Full public access |
| DOI | Sandbox DOI | Permanent DOI |
| Recommended for | First submissions, testing | Final datasets |
Auto-publish (-a, --auto-publish)
- Description: Automatically publish the record after submission
- Behavior:
- Without
-a: Record created in draft state (you can review before publishing) - With
-a: Record immediately published and public
- Without
- Usage:
-aor--auto-publish(no value needed) - Recommendation:
- Omit for first submission (review before publishing)
- Use
-afor batch submissions after testing
Data Selection (-d, --data-selection)
-
Description: Controls which NMR data types are shown in NMRium
-
Choices:
ft: Show only processed (Fourier Transform) spectra (default)fid: Show only raw FID databoth: Show both processed and FIDpreferFT: If both FID and processed are available, show processedpreferFID: If both FID and processed are available, show FID
-
Default:
ft -
Usage:
-d ft,--data-selection both -
Example: For comparing raw and processed data:
-d both
Log Level (-l, --log-level)
-
Description: Sets the verbosity level of the logger output
-
Choices:
trace: Most verbose; logs detailed execution trace informationdebug: Logs debugging information and variable valuesinfo: Standard logging level (recommended); logs important information (default)warn: Logs warnings and errors onlyerror: Logs errors onlyfatal: Logs only fatal errorssilent: No output at all
-
Default:
info -
Usage:
-l debug,--log-level warn -
Example: For detailed debugging:
-l debugor-l trace -
Recommendation:
- Use
info(default) for normal submissions - Use
debugortracewhen troubleshooting issues - Use
warnorsilentfor batch operations to reduce output
- Use
Visit the NMRium dataset on Zenodo to browse submitted datasets.